Jiong Zhang
Boosting Overlapping Organoid Instance Segmentation Using Pseudo-Label Unmixing and Synthesis-Assisted Learning
Jan 10, 2026Abstract:Organoids, sophisticated in vitro models of human tissues, are crucial for medical research due to their ability to simulate organ functions and assess drug responses accurately. Accurate organoid instance segmentation is critical for quantifying their dynamic behaviors, yet remains profoundly limited by high-quality annotated datasets and pervasive overlap in microscopy imaging. While semi-supervised learning (SSL) offers a solution to alleviate reliance on scarce labeled data, conventional SSL frameworks suffer from biases induced by noisy pseudo-labels, particularly in overlapping regions. Synthesis-assisted SSL (SA-SSL) has been proposed for mitigating training biases in semi-supervised semantic segmentation. We present the first adaptation of SA-SSL to organoid instance segmentation and reveal that SA-SSL struggles to disentangle intertwined organoids, often misrepresenting overlapping instances as a single entity. To overcome this, we propose Pseudo-Label Unmixing (PLU), which identifies erroneous pseudo-labels for overlapping instances and then regenerates organoid labels through instance decomposition. For image synthesis, we apply a contour-based approach to synthesize organoid instances efficiently, particularly for overlapping cases. Instance-level augmentations (IA) on pseudo-labels before image synthesis further enhances the effect of synthetic data (SD). Rigorous experiments on two organoid datasets demonstrate our method's effectiveness, achieving performance comparable to fully supervised models using only 10% labeled data, and state-of-the-art results. Ablation studies validate the contributions of PLU, contour-based synthesis, and augmentation-aware training. By addressing overlap at both pseudo-label and synthesis levels, our work advances scalable, label-efficient organoid analysis, unlocking new potential for high-throughput applications in precision medicine.
CLIP-DR: Textual Knowledge-Guided Diabetic Retinopathy Grading with Ranking-aware Prompting
Jul 04, 2024Abstract:Diabetic retinopathy (DR) is a complication of diabetes and usually takes decades to reach sight-threatening levels. Accurate and robust detection of DR severity is critical for the timely management and treatment of diabetes. However, most current DR grading methods suffer from insufficient robustness to data variability (\textit{e.g.} colour fundus images), posing a significant difficulty for accurate and robust grading. In this work, we propose a novel DR grading framework CLIP-DR based on three observations: 1) Recent pre-trained visual language models, such as CLIP, showcase a notable capacity for generalisation across various downstream tasks, serving as effective baseline models. 2) The grading of image-text pairs for DR often adheres to a discernible natural sequence, yet most existing DR grading methods have primarily overlooked this aspect. 3) A long-tailed distribution among DR severity levels complicates the grading process. This work proposes a novel ranking-aware prompting strategy to help the CLIP model exploit the ordinal information. Specifically, we sequentially design learnable prompts between neighbouring text-image pairs in two different ranking directions. Additionally, we introduce a Similarity Matrix Smooth module into the structure of CLIP to balance the class distribution. Finally, we perform extensive comparisons with several state-of-the-art methods on the GDRBench benchmark, demonstrating our CLIP-DR's robustness and superior performance. The implementation code is available \footnote{\url{https://github.com/Qinkaiyu/CLIP-DR}
GAPNet: Granularity Attention Network with Anatomy-Prior-Constraint for Carotid Artery Segmentation
Jun 27, 2024



Abstract:Atherosclerosis is a chronic, progressive disease that primarily affects the arterial walls. It is one of the major causes of cardiovascular disease. Magnetic Resonance (MR) black-blood vessel wall imaging (BB-VWI) offers crucial insights into vascular disease diagnosis by clearly visualizing vascular structures. However, the complex anatomy of the neck poses challenges in distinguishing the carotid artery (CA) from surrounding structures, especially with changes like atherosclerosis. In order to address these issues, we propose GAPNet, which is a consisting of a novel geometric prior deduced from.
DSCA: A Digital Subtraction Angiography Sequence Dataset and Spatio-Temporal Model for Cerebral Artery Segmentation
Jun 01, 2024



Abstract:Cerebrovascular diseases (CVDs) remain a leading cause of global disability and mortality. Digital Subtraction Angiography (DSA) sequences, recognized as the golden standard for diagnosing CVDs, can clearly visualize the dynamic flow and reveal pathological conditions within the cerebrovasculature. Therefore, precise segmentation of cerebral arteries (CAs) and classification between their main trunks and branches are crucial for physicians to accurately quantify diseases. However, achieving accurate CA segmentation in DSA sequences remains a challenging task due to small vessels with low contrast, and ambiguity between vessels and residual skull structures. Moreover, the lack of publicly available datasets limits exploration in the field. In this paper, we introduce a DSA Sequence-based Cerebral Artery segmentation dataset (DSCA), the first publicly accessible dataset designed specifically for pixel-level semantic segmentation of CAs. Additionally, we propose DSANet, a spatio-temporal network for CA segmentation in DSA sequences. Unlike existing DSA segmentation methods that focus only on a single frame, the proposed DSANet introduces a separate temporal encoding branch to capture dynamic vessel details across multiple frames. To enhance small vessel segmentation and improve vessel connectivity, we design a novel TemporalFormer module to capture global context and correlations among sequential frames. Furthermore, we develop a Spatio-Temporal Fusion (STF) module to effectively integrate spatial and temporal features from the encoder. Extensive experiments demonstrate that DSANet outperforms other state-of-the-art methods in CA segmentation, achieving a Dice of 0.9033.
Automating Vessel Segmentation in the Heart and Brain: A Trend to Develop Multi-Modality and Label-Efficient Deep Learning Techniques
Apr 02, 2024



Abstract:Cardio-cerebrovascular diseases are the leading causes of mortality worldwide, whose accurate blood vessel segmentation is significant for both scientific research and clinical usage. However, segmenting cardio-cerebrovascular structures from medical images is very challenging due to the presence of thin or blurred vascular shapes, imbalanced distribution of vessel and non-vessel pixels, and interference from imaging artifacts. These difficulties make manual or semi-manual segmentation methods highly time-consuming, labor-intensive, and prone to errors with interobserver variability, where different experts may produce different segmentations from a variety of modalities. Consequently, there is a growing interest in developing automated algorithms. This paper provides an up-to-date survey of deep learning techniques, for cardio-cerebrovascular segmentation. It analyzes the research landscape, surveys recent approaches, and discusses challenges such as the scarcity of accurately annotated data and variability. This paper also illustrates the urgent needs for developing multi-modality label-efficient deep learning techniques. To the best of our knowledge, this paper is the first comprehensive survey of deep learning approaches that effectively segment vessels in both the heart and brain. It aims to advance automated segmentation techniques for cardio-cerebrovascular diseases, benefiting researchers and healthcare professionals.
PEFA: Parameter-Free Adapters for Large-scale Embedding-based Retrieval Models
Dec 06, 2023Abstract:Embedding-based Retrieval Models (ERMs) have emerged as a promising framework for large-scale text retrieval problems due to powerful large language models. Nevertheless, fine-tuning ERMs to reach state-of-the-art results can be expensive due to the extreme scale of data as well as the complexity of multi-stages pipelines (e.g., pre-training, fine-tuning, distillation). In this work, we propose the PEFA framework, namely ParamEter-Free Adapters, for fast tuning of ERMs without any backward pass in the optimization. At index building stage, PEFA equips the ERM with a non-parametric k-nearest neighbor (kNN) component. At inference stage, PEFA performs a convex combination of two scoring functions, one from the ERM and the other from the kNN. Based on the neighborhood definition, PEFA framework induces two realizations, namely PEFA-XL (i.e., extra large) using double ANN indices and PEFA-XS (i.e., extra small) using a single ANN index. Empirically, PEFA achieves significant improvement on two retrieval applications. For document retrieval, regarding Recall@100 metric, PEFA improves not only pre-trained ERMs on Trivia-QA by an average of 13.2%, but also fine-tuned ERMs on NQ-320K by an average of 5.5%, respectively. For product search, PEFA improves the Recall@100 of the fine-tuned ERMs by an average of 5.3% and 14.5%, for PEFA-XS and PEFA-XL, respectively. Our code is available at https://github.com/amzn/pecos/tree/mainline/examples/pefa-wsdm24.
Polar-Net: A Clinical-Friendly Model for Alzheimer's Disease Detection in OCTA Images
Nov 10, 2023Abstract:Optical Coherence Tomography Angiography (OCTA) is a promising tool for detecting Alzheimer's disease (AD) by imaging the retinal microvasculature. Ophthalmologists commonly use region-based analysis, such as the ETDRS grid, to study OCTA image biomarkers and understand the correlation with AD. However, existing studies have used general deep computer vision methods, which present challenges in providing interpretable results and leveraging clinical prior knowledge. To address these challenges, we propose a novel deep-learning framework called Polar-Net. Our approach involves mapping OCTA images from Cartesian coordinates to polar coordinates, which allows for the use of approximate sector convolution and enables the implementation of the ETDRS grid-based regional analysis method commonly used in clinical practice. Furthermore, Polar-Net incorporates clinical prior information of each sector region into the training process, which further enhances its performance. Additionally, our framework adapts to acquire the importance of the corresponding retinal region, which helps researchers and clinicians understand the model's decision-making process in detecting AD and assess its conformity to clinical observations. Through evaluations on private and public datasets, we have demonstrated that Polar-Net outperforms existing state-of-the-art methods and provides more valuable pathological evidence for the association between retinal vascular changes and AD. In addition, we also show that the two innovative modules introduced in our framework have a significant impact on improving overall performance.
Representer Point Selection for Explaining Regularized High-dimensional Models
May 31, 2023



Abstract:We introduce a novel class of sample-based explanations we term high-dimensional representers, that can be used to explain the predictions of a regularized high-dimensional model in terms of importance weights for each of the training samples. Our workhorse is a novel representer theorem for general regularized high-dimensional models, which decomposes the model prediction in terms of contributions from each of the training samples: with positive (negative) values corresponding to positive (negative) impact training samples to the model's prediction. We derive consequences for the canonical instances of $\ell_1$ regularized sparse models, and nuclear norm regularized low-rank models. As a case study, we further investigate the application of low-rank models in the context of collaborative filtering, where we instantiate high-dimensional representers for specific popular classes of models. Finally, we study the empirical performance of our proposed methods on three real-world binary classification datasets and two recommender system datasets. We also showcase the utility of high-dimensional representers in explaining model recommendations.
PINA: Leveraging Side Information in eXtreme Multi-label Classification via Predicted Instance Neighborhood Aggregation
May 21, 2023Abstract:The eXtreme Multi-label Classification~(XMC) problem seeks to find relevant labels from an exceptionally large label space. Most of the existing XMC learners focus on the extraction of semantic features from input query text. However, conventional XMC studies usually neglect the side information of instances and labels, which can be of use in many real-world applications such as recommendation systems and e-commerce product search. We propose Predicted Instance Neighborhood Aggregation (PINA), a data enhancement method for the general XMC problem that leverages beneficial side information. Unlike most existing XMC frameworks that treat labels and input instances as featureless indicators and independent entries, PINA extracts information from the label metadata and the correlations among training instances. Extensive experimental results demonstrate the consistent gain of PINA on various XMC tasks compared to the state-of-the-art methods: PINA offers a gain in accuracy compared to standard XR-Transformers on five public benchmark datasets. Moreover, PINA achieves a $\sim 5\%$ gain in accuracy on the largest dataset LF-AmazonTitles-1.3M. Our implementation is publicly available.
Retinal Structure Detection in OCTA Image via Voting-based Multi-task Learning
Aug 23, 2022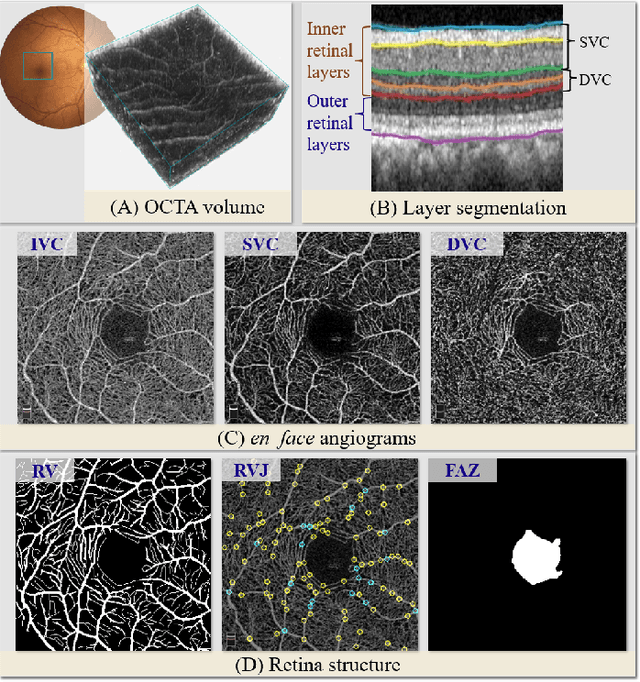
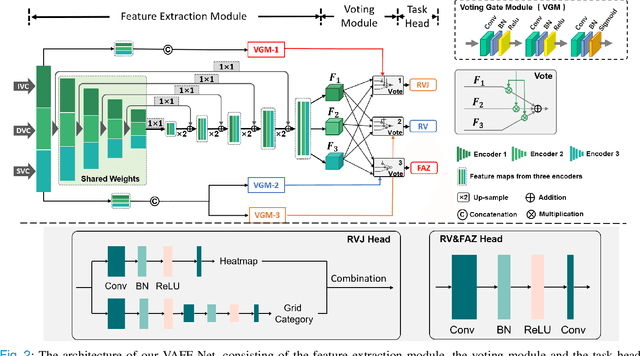
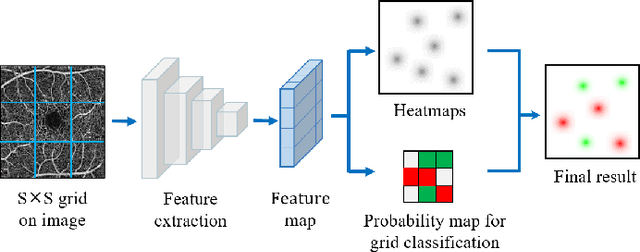
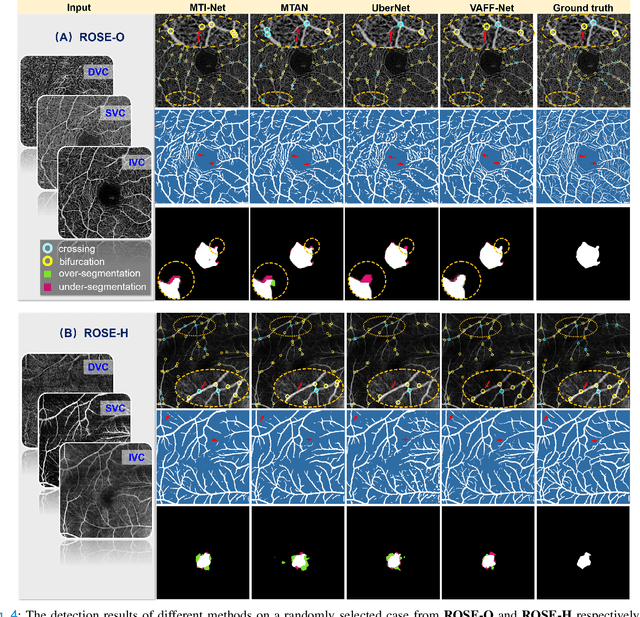
Abstract:Automated detection of retinal structures, such as retinal vessels (RV), the foveal avascular zone (FAZ), and retinal vascular junctions (RVJ), are of great importance for understanding diseases of the eye and clinical decision-making. In this paper, we propose a novel Voting-based Adaptive Feature Fusion multi-task network (VAFF-Net) for joint segmentation, detection, and classification of RV, FAZ, and RVJ in optical coherence tomography angiography (OCTA). A task-specific voting gate module is proposed to adaptively extract and fuse different features for specific tasks at two levels: features at different spatial positions from a single encoder, and features from multiple encoders. In particular, since the complexity of the microvasculature in OCTA images makes simultaneous precise localization and classification of retinal vascular junctions into bifurcation/crossing a challenging task, we specifically design a task head by combining the heatmap regression and grid classification. We take advantage of three different \textit{en face} angiograms from various retinal layers, rather than following existing methods that use only a single \textit{en face}. To facilitate further research, part of these datasets with the source code and evaluation benchmark have been released for public access:https://github.com/iMED-Lab/VAFF-Net.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge